
Systems biology: from data mining, through network inference to pathway modelling.
Expositor: Gustavo Stolovitzky, Watson Research Center, IBM, New York.
Colaborador: Diego Fernandez Slezak.
The nascent field of systems biology shares with the more traditional field of physiology the aim of understanding how biological systems work. However, it differentiates from more traditional physiology on two counts: its quantitative thrust and the availability of high throughput data of unprecedented amount and quality. This mini-course will introduce the main thrusts of today’s research in systems biology. The course will emphasize the role that physicists can play in this active field of research. It will be divided in three units as follows (see figure).
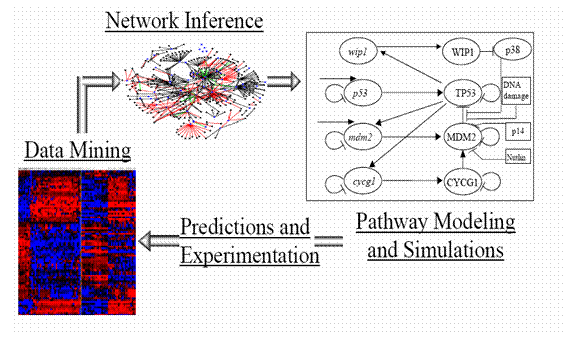
- Data mining (2 hours): We will first discuss a few of the many novel technologies that allow researchers to probe the state of a cell at the transcript, protein and metabolic level. We will then concentrate on transcription data, and discuss some of the noise associated with this technologies [1,2] and review the more traditional methods to infer genes that are associated with a particular type of disease or cellular state [3].
- Network Inference (2 hours): One of the present challenges in biological research is the organization of the data originating from high-throughput technologies such as transcriptional profiling. One way in which this information can be organized is in the form of networks of influences, physical or statistical, between genes. These interactions provide a high level overview of how genes work together to achieve a specific function within the cellular environment. I will present some strategies and methods for the reconstruction of gene regulatory networks from gene expression data, and analyze the source of possible reconstruction errors due to noise, non-linearity, multiple inputs to the target nodes, etc. I will exemplify these approaches in the context of the gene regulatory network, E. coli network [4] and human B-cells [5]. We will also discuss how to mine these networks using graph theoretic methods to extract valuable biological information in the context of a recently studied cell signalling network of hippocampal neurons [6].
- Pathway modelling (2 hours). Global network reconstruction is only a step to the creation of predictive models with mechanistic detail which is ultimately the goal of systems biology [7]. We are only beginning to have enough data to construct rather rudimentary models and simulations of a handful of cellular processes. Typically, present models are parsimonious descriptions containing mainly key players with speculative features to fill in gaps in the current knowledge. We will exemplify the present state of mechanistic modelling of important pathways in the context of then NFkB pathway [8] and the p53 pathway [9].
1. Tu Y, Stolovitzky G, Klein U (2002) Quantitative noise analysis for gene expression microarray experiments. Proc Natl Acad Sci U S A 99: 14031-14036.
2. Stolovitzky GA, Kundaje A, Held GA, Duggar KH, Haudenschild CD, et al. (2005) Statistical analysis of MPSS measurements: Application to the study of LPS-activated macrophage gene expression. Proc Natl Acad Sci U S A.
3. Stolovitzky G (2003) Gene selection in microarray data: the elephant, the blind men and our algorithms. Curr Opin Struct Biol 13: 370-376.
4. Rice JJ, Tu Y, Stolovitzky G (2005) Reconstructing biological networks using conditional correlation analysis. Bioinformatics 21: 765-773. Epub 2004 Oct 2014.
5. Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, et al. (2005) Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382-390. Epub 2005 Mar 2020.
6. Ma'ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, et al. (2005) Formation of regulatory patterns during signal propagation in a Mammalian cellular network. Science 309: 1078-1083.
7. Rice JJ, Stolovitzky G (2004) Making the most of it: pathway reconstruction and integrative simulation using the data at hand. Biosilico 2: 70-77.
8. Hoffmann A, Levchenko A, Scott ML, Baltimore D (2002) The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298: 1241-1245.
9. Ma L, J. W, Rice JJ, Hu W, Levine AJ, et al. (2005) A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci U S A 102, 1402.